Computational biologist Janani Ravi, PhD, assistant professor of biomedical informatics at the University of Colorado School of Medicine, is looking deep into the functions of bacterial proteins and helping others do the same.
“We don’t always know where the biology is going to take us, but we do know what kinds of computational methods will help us get there. We’ve taken our successes and used them as a model to continue to help other researchers, too,” says Janani, who this month published a paper in the American Society for Microbiology journal mSystems detailing the evolution of the phage shock protein (PSP) system.
In the paper, Janani and fellow researchers explain how they discovered that the core protein of the bacterial PSP system was actually present in the last universal common ancestor, an organism that lived billions of years ago and from which all life descended. Since then, that original system has evolved into the many distinct PSP systems seen across life today, from microbes like bacteria and archaea, to eukaryotes, including yeasts, plants, and animals.
The discovery also led to the JRaviLab creating a web application that enables other researchers to visualize the features and evolution of any protein of interest in its contexts across the tree of life.
Making sense of the microbial genome
Researchers over the past few years have independently discovered PSP systems, noting that there seemed to be a main protein, but partner proteins were all very different despite performing similar functions.
“Despite this divergence, in each case, the PSP system was protecting the cell membrane from any external stress,” Janani explains.
For Janani, this was intriguing. She wanted to know which were the main proteins, which were the variable partners, and how they came together to produce a conserved stress response function. With partner collaborators at Rutgers and the National Institutes of Health, an expansive study was underway to unravel the mystery.
Looking at approximately 7,000 species across the tree of life — including bacteria, archaea, viruses, and eukaryotes — the researchers concluded that the main protein in the PSP system can be traced all the way back to one common ancestor.
“This ancient ancestor seems to have had a very similar protein to what we’re seeing today,” Janani explains. “And its existence tells us how critical this protein is in responding to stress and maintaining that membrane homeostasis all across life.”
“We then realized there were multiple central proteins, not just one,” she continues. “And PSP systems seem to have a life of their own. There are different partners based on which organisms it’s part of, similar to the first protein we studied.”
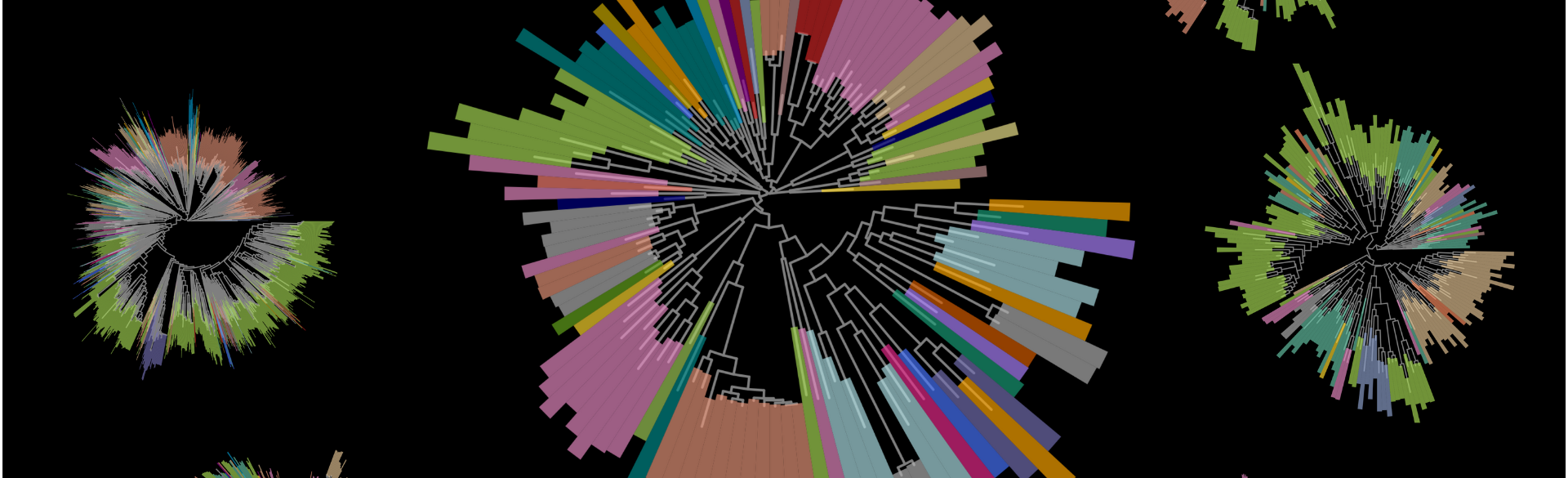
![psp_cover_art_blk[66949]](https://news.cuanschutz.edu/hs-fs/hubfs/Department%20of%20Biomedical%20Infomatics/psp_cover_art_blk%5B66949%5D.png?width=475&height=447&name=psp_cover_art_blk%5B66949%5D.png)
The cover art for the paper published in mSystems features the phyletic spread of the key effector protein PspA (center) and a few of its many PSP partners. Image courtesy of Raymond Lesiyon, Evan Brenner, and Janani Ravi.
Suddenly, Janani and the researchers were seeing these central proteins everywhere.
“Our method of breaking down the proteins into their homologues, meaning other closely related proteins across the tree of life, then breaking them down further into these functional protein domain subunits, was extremely helpful in uncovering new biology,” Janani says.
Broadening accessibility
With so much success in their recent work, Janani’s team built MolEvolvR, a novel web application that allows researchers to do what they have done but in their own specialties and research projects.
“We wanted to make this method available for biologists who don’t have the same computational experience as we have. We thought, ‘Why not just build an accessible application?’ So, we did,” Janani says.
Now, scientists around the world are beginning to use MolEvolvR for their own research.
“We’re hearing a lot of excitement from collaborators and other users,” Janani says. “We hope this takes them into new discovery spaces where they may not have previously thought possible. Maybe they thought a particular protein was unique, but with our application, they can see if that’s true. This will help aid the discovery of more protein systems, and understanding what proteins do and which species have them helping us understand what’s driving evolution and diversity of life.”


.png)
